Lesson 3 - Rtracklayer, Biomart a GenomicRanges
Pokracujeme v praci s anotacnymi datami, tentokrat ale priamo z R. Zacneme niekolkymi beznymi sposobmi ako ziskat data pre dalsiu pracu:
Navstivte https://www.arabidopsis.org/download/index-auto.jsp?dir=/download_files/Genes/TAIR10_genome_release a ziskajte sekvencie chromozomov Arabidopsis thaliana, verziu genomu TAIR10 a anotacne subory v GFF3 formate pre geny a transpozony.
Nacitame genom (sekvencie chromozomov v multiFASTA formate), ktory budeme anotovat:
library(Biostrings)
genome_seq <- readDNAStringSet("TAIR10.fas")
genome_seq## A DNAStringSet instance of length 7
## width seq names
## [1] 30427671 CCCTAAACCCTAAACCCTAAACCCTAAACCTCTGAATCCTTAATCC...TTGATTAGGGTTTAGGGTTTAGGGTTTAGGGTTTAGGGTTTAGGG Chr1 CHROMOSOME d...
## [2] 19698289 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN...GGGTTTAGGGTTTAGGGTTTAGGGTTTAGGGTTTAGGGTTTAGGG Chr2 CHROMOSOME d...
## [3] 23459830 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN...TTCCTCTACTTCTACCCTAAACCCTAAACCCTAAACCCTAAACCC Chr3 CHROMOSOME d...
## [4] 18585056 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN...AGGGTTTAGGGTTAAGGGTTAAGGGTTTAGGGTTTAGGGTTTAGG Chr4 CHROMOSOME d...
## [5] 26975502 TATACCATGTACCCTCAACCTTAAAACCCTAAAACCTATACTATAA...GGGTATGGTATAGGTATATGGTTTAGGATTTAGGGTTTTTAGATC Chr5 CHROMOSOME d...
## [6] 154478 ATGGGCGAACGACGGGAATTGAACCCGCGATGGTGAATTCACAATC...TACAAAGGCGGAAAAAGAAATCATAATAACTTGGTCCCGGGCATC chloroplast CHROM...
## [7] 366924 GGATCCGTTCGAAACAGGTTAGCCTACTATAATATAAGGATTGGAT...AGGAGAATGCATCATTCTTATCGCAGAATGGAAACAAACCGGATT mitochondria CHRO...Nacitame anotacne sekvencie:
library(rtracklayer)
genome_gff <- import.gff3("TAIR10_GFF3_genes_transposons.gff")
genome_gff## GRanges object with 656309 ranges and 11 metadata columns:
## seqnames ranges strand | source type score phase ID
## <Rle> <IRanges> <Rle> | <factor> <factor> <numeric> <integer> <character>
## [1] Chr1 1-30427671 * | TAIR10 chromosome <NA> <NA> Chr1
## [2] Chr1 3631-5899 + | TAIR10 gene <NA> <NA> AT1G01010
## [3] Chr1 3631-5899 + | TAIR10 mRNA <NA> <NA> AT1G01010.1
## [4] Chr1 3760-5630 + | TAIR10 protein <NA> <NA> AT1G01010.1-Protein
## [5] Chr1 3631-3913 + | TAIR10 exon <NA> <NA> <NA>
## ... ... ... ... . ... ... ... ... ...
## [656305] Chr5 26893818-26893895 - | TAIR10 transposon_fragment <NA> <NA> <NA>
## [656306] Chr5 26925227-26925333 + | TAIR10 transposable_element <NA> <NA> AT5TE97000
## [656307] Chr5 26925227-26925333 + | TAIR10 transposon_fragment <NA> <NA> <NA>
## [656308] Chr5 26965908-26966653 - | TAIR10 transposable_element <NA> <NA> AT5TE97150
## [656309] Chr5 26965908-26966653 - | TAIR10 transposon_fragment <NA> <NA> <NA>
## Name Note Parent Index Derives_from Alias
## <character> <CharacterList> <CharacterList> <character> <character> <CharacterList>
## [1] Chr1 <NA> <NA>
## [2] AT1G01010 protein_coding_gene <NA> <NA>
## [3] AT1G01010.1 AT1G01010 1 <NA>
## [4] AT1G01010.1 <NA> AT1G01010.1
## [5] <NA> AT1G01010.1 <NA> <NA>
## ... ... ... ... ... ... ...
## [656305] <NA> AT5TE96885 <NA> <NA>
## [656306] AT5TE97000 <NA> <NA> ATHILA8B
## [656307] <NA> AT5TE97000 <NA> <NA>
## [656308] AT5TE97150 <NA> <NA> ATREP9
## [656309] <NA> AT5TE97150 <NA> <NA>
## -------
## seqinfo: 7 sequences from an unspecified genome; no seqlengthsAk sa jedna o genom, mozeme sekvenciu ziskat instalaciou prislusneho balicka v R:
# if (!requireNamespace("BiocManager", quietly = TRUE))
# install.packages("BiocManager")
#
# BiocManager::install("BSgenome.Athaliana.TAIR.TAIR9")a jeho zavolanim:
library(BSgenome.Athaliana.TAIR.TAIR9)
BSgenome.Athaliana.TAIR.TAIR9## Arabidopsis genome:
## # organism: Arabidopsis thaliana (Arabidopsis)
## # provider: TAIR
## # provider version: TAIR9
## # release date: June 9, 2009
## # release name: TAIR9 Genome Release
## # 7 sequences:
## # Chr1 Chr2 Chr3 Chr4 Chr5 ChrM ChrC
## # (use 'seqnames()' to see all the sequence names, use the '$' or '[[' operator to access a given sequence)BSgenome.Athaliana.TAIR.TAIR9$Chr1## 30427671-letter "DNAString" instance
## seq: CCCTAAACCCTAAACCCTAAACCCTAAACCTCTGAATCCTTAATCCCTAAATCCCTAAAT...ATTAGTTACCATGTCTTGATTAGGGTTTAGGGTTTAGGGTTTAGGGTTTAGGGTTTAGGGAnotacie v R/Bioconductor su k dispozicii ako balicky org.*.db, v tomto pripade org.At.tair.db:
library(org.At.tair.db)
columns(org.At.tair.db)## [1] "ARACYC" "ARACYCENZYME" "ENTREZID" "ENZYME" "EVIDENCE" "EVIDENCEALL" "GENENAME" "GO"
## [9] "GOALL" "ONTOLOGY" "ONTOLOGYALL" "PATH" "PMID" "REFSEQ" "SYMBOL" "TAIR"symbols <- c('ARR1', 'ARR2', 'ARR22', 'PHYA', 'PHYB')
mapIds(org.At.tair.db, symbols, 'TAIR', 'SYMBOL')## 'select()' returned 1:1 mapping between keys and columns## ARR1 ARR2 ARR22 PHYA PHYB
## "AT3G16857" "AT4G16110" "AT3G04280" "AT1G09570" "AT2G18790"Vizualizacia sa v sucasnosti casto robi pomocou balicka ggbio:
ggbio::autoplot(genome_gff)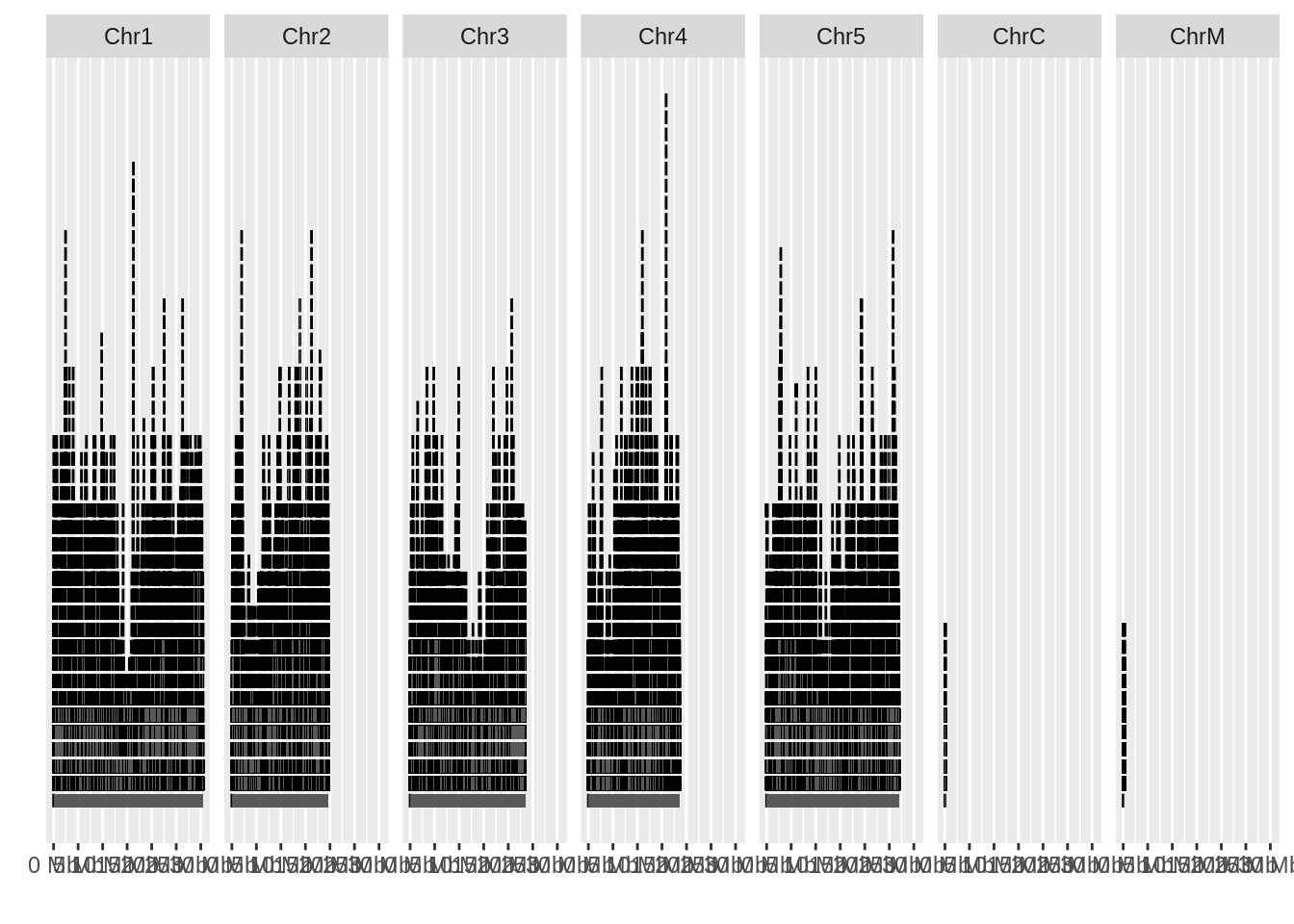
Dalsim zaujimavym zdrojom dat je system Biomart (distribuovany datovy sklad):
library("biomaRt")
listMarts()## biomart version
## 1 ENSEMBL_MART_ENSEMBL Ensembl Genes 99
## 2 ENSEMBL_MART_MOUSE Mouse strains 99
## 3 ENSEMBL_MART_SNP Ensembl Variation 99
## 4 ENSEMBL_MART_FUNCGEN Ensembl Regulation 99ensembl <- useMart("ensembl")
datasets <- listDatasets(ensembl)
head(datasets)## dataset description version
## 1 acalliptera_gene_ensembl Eastern happy genes (fAstCal1.2) fAstCal1.2
## 2 acarolinensis_gene_ensembl Anole lizard genes (AnoCar2.0) AnoCar2.0
## 3 acchrysaetos_gene_ensembl Golden eagle genes (bAquChr1.2) bAquChr1.2
## 4 acitrinellus_gene_ensembl Midas cichlid genes (Midas_v5) Midas_v5
## 5 amelanoleuca_gene_ensembl Panda genes (ailMel1) ailMel1
## 6 amexicanus_gene_ensembl Mexican tetra genes (Astyanax_mexicanus-2.0) Astyanax_mexicanus-2.0ensembl = useMart("ensembl",dataset="hsapiens_gene_ensembl")
filters = listFilters(ensembl)
filters[1:5,]## name description
## 1 chromosome_name Chromosome/scaffold name
## 2 start Start
## 3 end End
## 4 band_start Band Start
## 5 band_end Band Endattributes = listAttributes(ensembl)
attributes[1:5,]## name description page
## 1 ensembl_gene_id Gene stable ID feature_page
## 2 ensembl_gene_id_version Gene stable ID version feature_page
## 3 ensembl_transcript_id Transcript stable ID feature_page
## 4 ensembl_transcript_id_version Transcript stable ID version feature_page
## 5 ensembl_peptide_id Protein stable ID feature_pageaffyids=c("202763_at","209310_s_at","207500_at")
getBM(attributes=c('affy_hg_u133_plus_2', 'entrezgene_id'),
filters = 'affy_hg_u133_plus_2',
values = affyids,
mart = ensembl)## affy_hg_u133_plus_2 entrezgene_id
## 1 209310_s_at 837
## 2 207500_at 838
## 3 202763_at 836